Research
"The overall aim of PROHITS is to integrate experimental and computational approaches that can resolve the proteome of prokaryotes at single cell level, and at a range of temperatures, in order to improve our understanding of thermophile biology and to guide the optimized design of cell factories"
Introduction
The increasingly urgent need for a sustainable climate and ecosystem friendly economy demands the development of biobased alternatives to the petrochemical industry. "Cell factories", in which organisms convert renewable biomass into various biochemicals and -materials that would otherwise be derived from fossil fuels or harsh chemistry, are the solution.
Currently cell factories are typically based on commonly used micro- organisms such as E. coli, which are limited to low temperatures. This prevents cost-effective bioconversions, which in turn hampers industrial-scale viability in competition with fossil fuels. Industry focus is therefore shifting towards the use of thermophilic prokaryotes (bacteria and archaea) that operate at high temperatures, enabling more efficient production processes, for example by combining fermentation with high temperature biomass pre- treatment and by lowering contamination risks.
However, we know very little about the proteome of thermophiles, their cellular protein machinery. Obtaining this knowledge is essential for the creation of successful cell factories:
- Proteins are the molecular machines that drive cell factories; they transform basic compounds into molecular building blocks. To engineer thermophiles for improved production, we have to understand their proteome so that we can adapt thermophiles to make the desired product in the most efficient way;
- The bioreactors in which thermophiles are grown can have large variations in temperature; we need to know how these variations affect the proteome (and function) of these thermophiles in order to optimise production.
While DNA (genomics) and RNA (transcriptomics) knowledge has already revolutionised our understanding of biology and medicine, very little is known about the way cells maintain their proteome, and how it can dynamically change.
The next big step in our "-omics" based understanding of life is therefore mass spectrometry (MS)-driven proteomics, which enables study of the in vivo proteome of cells. Specifically, we need to understand how the proteome of thermophiles responds to temperature - for example, which proteins unfold or aggregate, in which order?
Additionally, each cell behaves differently, therefore the ability to study the proteome of individual cells can give us insights in the various ways cells respond to environmental triggers, allowing us to select and expand specific cells, or cell combinations, for optimal bioreactor compatibility. In order to obtain these insights into thermophiles, advances in MS proteomics are required as current technology is insufficiently developed.The PROHITS doctoral network will advance
- microfluidics to separate single prokaryotic cells,
- mass spectrometry (MS) and its associated bioinformatics to handle single prokaryotic cells,
- MS-based thermal proteome profiling to study the thermal response of the proteome of thermophiles. We will use the information generated to
- understand the in vivo stability of proteins, and
- create genome-scale models of thermophiles to
- optimize them as cell factories.
The two main technological outcomes are:
- Instrumentation to determine the proteome of single prokaryotic cells and its response to temperature changes, with relevance for other fields (e.g. response of key prokaryotes to rising global temperatures).
- Improved cell factory modelling by including proteome and thermal response information, so guiding metabolic engineering approaches to optimize environmentally sustainable industrial processes.
The fundamental scientific insights we generate are to:
- Increase our general understanding of thermophilic proteomes, including (engineered) cell factories, at the resolution of single cells, thus capturing proteome variability at the finest level.
- Reveal the effect of temperature on thermophilic proteomes, down to the single protein level.
PROHITS achieves this by integrating dry and wet lab approaches, with a consortium that includes experts from both computational and experimental fields, covering microfluidics, mass spectrometry (MS), applied machine learning, structural bioinformatics, cell modelling, and microbial biotechnology.
The tight interaction between these approaches and fields provides the foundation to train 9 ESRs. These ESRs will be located in laboratories with leading scientific expertise in cell separation through microfluidics (Cellenion), MS proteomics (UD, CNRS, Bruker), proteome data analysis (VUB, VIB, UiB), proteome-level understanding of extremophiles (VUB, UD), single protein interpretation (VUB), and cell factories (UniVie, NovoArc).
PROHITS will integrate experiment and computation to create a pipeline applicable in both academic and industry settings, with the ESRs acquiring key translational and interdisciplinary skills in laboratories with extensive PhD training experience.
In addition, they will receive transversal and entrepreneurial skills training resulting in:
- Excellent understanding of both experiment and computation, and of the challenges and opportunities at the interface between these. This is particularly important as the reliable bioinformatics analysis of large-scale biological data an increasing bottleneck in academics and industry alike;
- The ability to solve challenges in the determination, (biological) interpretation, and (industrial) application of thermophilic proteomes, along with broader method applicability to all prokaryotes;
- Engagement with the societal impact of their research, and awareness of innovation opportunities.
Together, these comprehensive and complementary skills will enable the ESRs to constructively contribute to the European bioeconomy in both academic as well as industry settings. This ties into the long-term vision of the project to further develop the prototype pipeline within industry, and to provide continued training in proteomics analysis for (thermophilic) prokaryotes and cell factory development.
Work packages
Our specific objectives, each connected to a work package (WP), are to:
WP1: Enable single cell mass spectrometry for prokaryotes
-
Objectives:
- Separate single prokaryotic cells via microfluidics.
- Enable mass spectrometry instrumentation and computational pipelines to handle data for single prokaryotic cells.
- Requires: a combination of improved instrumentation for microfluidics (Cellenion) and mass spectrometry (CNRS, Bruker) with state-of-the-art academic knowledge on experimental (CNRS) and computational (VIB, UiB) approaches relevant for single-cell MS data, including visualization (UiB).
- Delivers: the capability to analyze the individual proteomes of prokaryotic cells, producing insights in proteome variability, also in response to environmental triggers (e.g. temperature).
WP2: Investigate the thermal response of thermophiles
-
Objectives:
- Ensure availability of thermophiles for project.
- Develop full pipeline for thermal proteome profiling (TPP) of thermophiles.
- Create library of thermophile TPP data.
- Visualize results for human interpretation.
- Requires: state-of-the-art academic knowledge on experimental (CNRS, UD) and computational (VIB, UiB) approaches relevant for thermal profiling, combined with biological insights in thermophiles (VUB) and including visualization (UiB).
- Delivers: the ability to investigate the thermal response of prokaryotic proteomes, with data produced for archaeal and bacterial thermophiles, with focus on organisms used to develop cell factories.
WP3: Improve our understanding of the proteome of cell factories, and adapt these accordingly
-
Objectives:
- Connect single protein information to the proteome (and back).
- Develop detailed and coarse genome-scale models (GEMs) for thermophiles.
- Improve existing cell factory.
- Pursue new alternatives for cell factories.
- Requires: existing cell factories and knowledge of industrial applications (NovoArc) combined with biological insights in cell factories (VUB), expertise in genome wide metabolic networks (GEMs) (UniVie) and connecting proteome level data to the individual protein level (VUB, VIB).
- Delivers: the tools to analyze and understand the proteomes of prokaryotic cells, for application to cell factories and to produce insights in the proteome response to environmental triggers (e.g. temperature).
WP4: Training and education
-
Objectives:
- Organising network-wide training activities, coordinating local training, supervision of research training.
- Ensuring ESR progress and career perspectives by personal career development plans (PCDPs).
- Monitoring and reporting in relation to training program.
WP5: Coordination and management
-
Objectives:
- Develop and sign the Consortium Agreement.
- Establish boards and committees.
- Recruit ESRs.
- Financial management of network.
- IP management.
- Data management.
WP6: Dissemination and communication
-
Objectives:
- Relate the project progress to the stakeholders and create an PROHITS-connected community.
- Involve ESRs in dissemination and communication as part of their training on transferable skills.
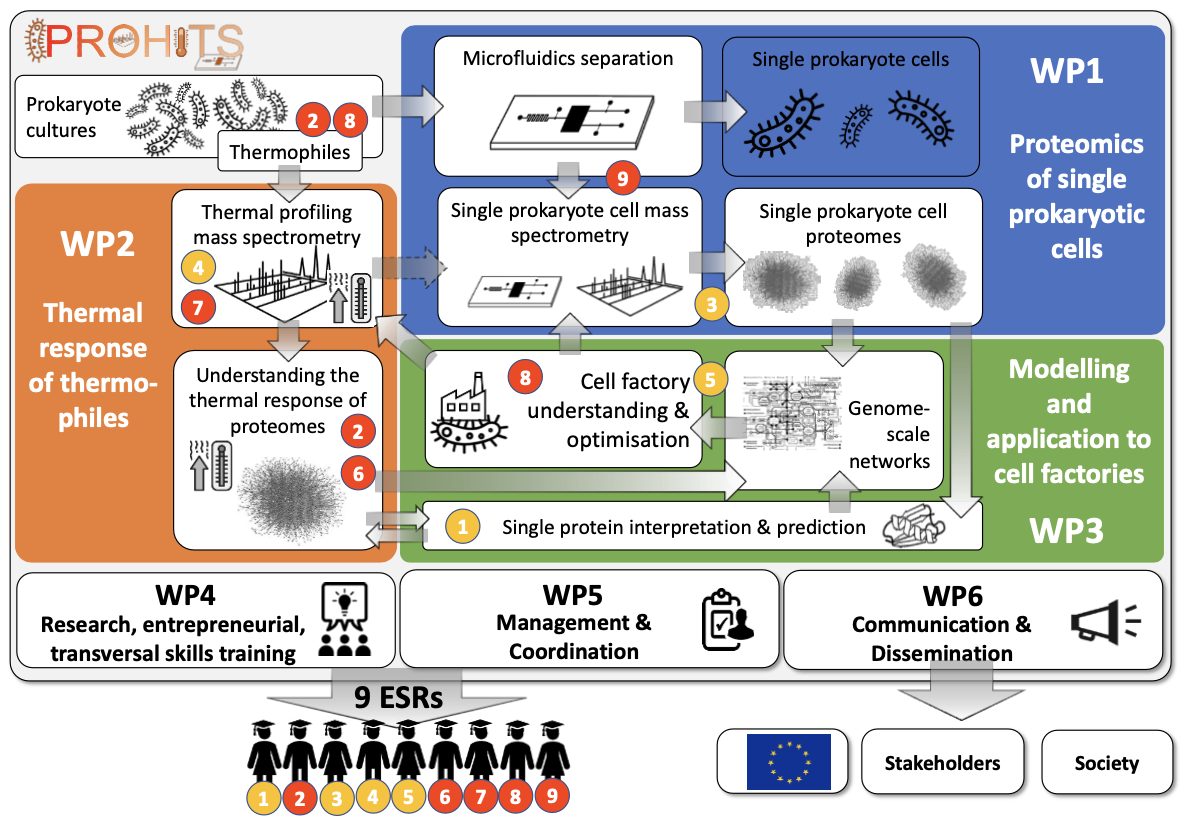
The main objectives are each connected to a work package (WP), with the scientific WPs tightly integrated with training, management and dissemination WPs (Figure 1.1a). The individual ESR projects are tailored to specific topics with the aim to create a pipeline from single cell prokaryote proteomics to cell factory understanding. The projects are interconnected through secondments, and offer multiple learning opportunities through other planned interactions with consortium partners. The close interaction between experiment and computation is increasingly crucial for competitive research: computational tools require experimental data and validation, while experimental results can be boosted considerably by cutting-edge computational approaches. We will therefore ensure that the ESRs develop the know-how, insight into and the optimal application of each approach in the context of their research, both through planned secondments, other planned interactions as well as informally through an ESR "buddy" system. The mix of industry and academics provides a wide range of environments.